Transcription factors are key regulators of gene expression,
playing a crucial role in determining which genes are turned on or off in a cell.
Predicting transcription factor binding sites in the genome is
a complex but essential task that helps researchers understand
how genes are regulated and how this regulation can impact various biological processes.
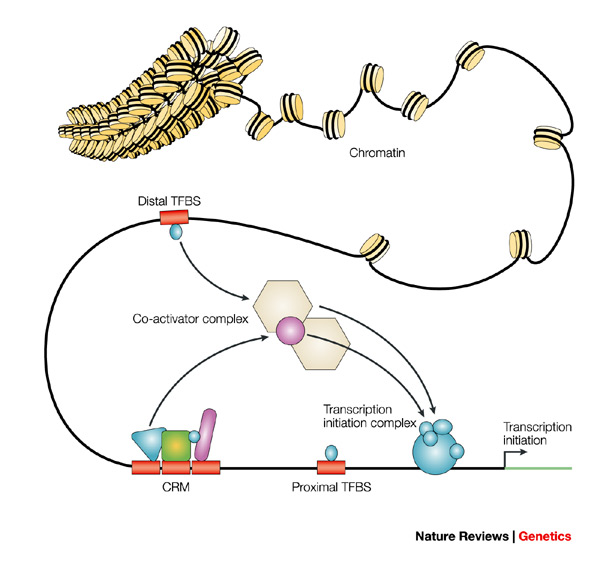
Transcription factor binding sites
Are specific DNA sequences where transcription factors bind to control gene expression. Predicting these sites involves identifying short DNA sequences that match known binding motifs for transcription factors.
This process is challenging due to the vast size and complexity of the genome, as well as the diverse range of transcription factors and their binding preferences.
Fig1: Transcription factor binding sites
Several computational methods have been developed to predict transcription factor binding sites.
These methods often rely on machine learning algorithms that analyze DNA sequences to identify patterns associated with transcription factor binding.
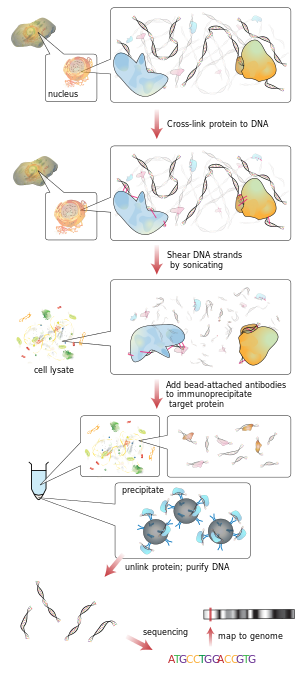
Additionally, experimental techniques such as chromatin immunoprecipitation sequencing (ChIP-seq) can be used to validate predicted binding sites and provide insights into the regulatory networks controlling gene expression.
Fig2: ChIP Sequencing